sangeranalyseR: simple and interactive processing of Sanger sequencing Data in R
Introduction
I developed sangeranalyseR while working as a research assistant at the Molecular Evolution and Phylogenetics Lab, led by Prof. Robert Lanfear during my exchange at the Australian National University. sangeranalyseR is now available on Bioconductor 3.12.
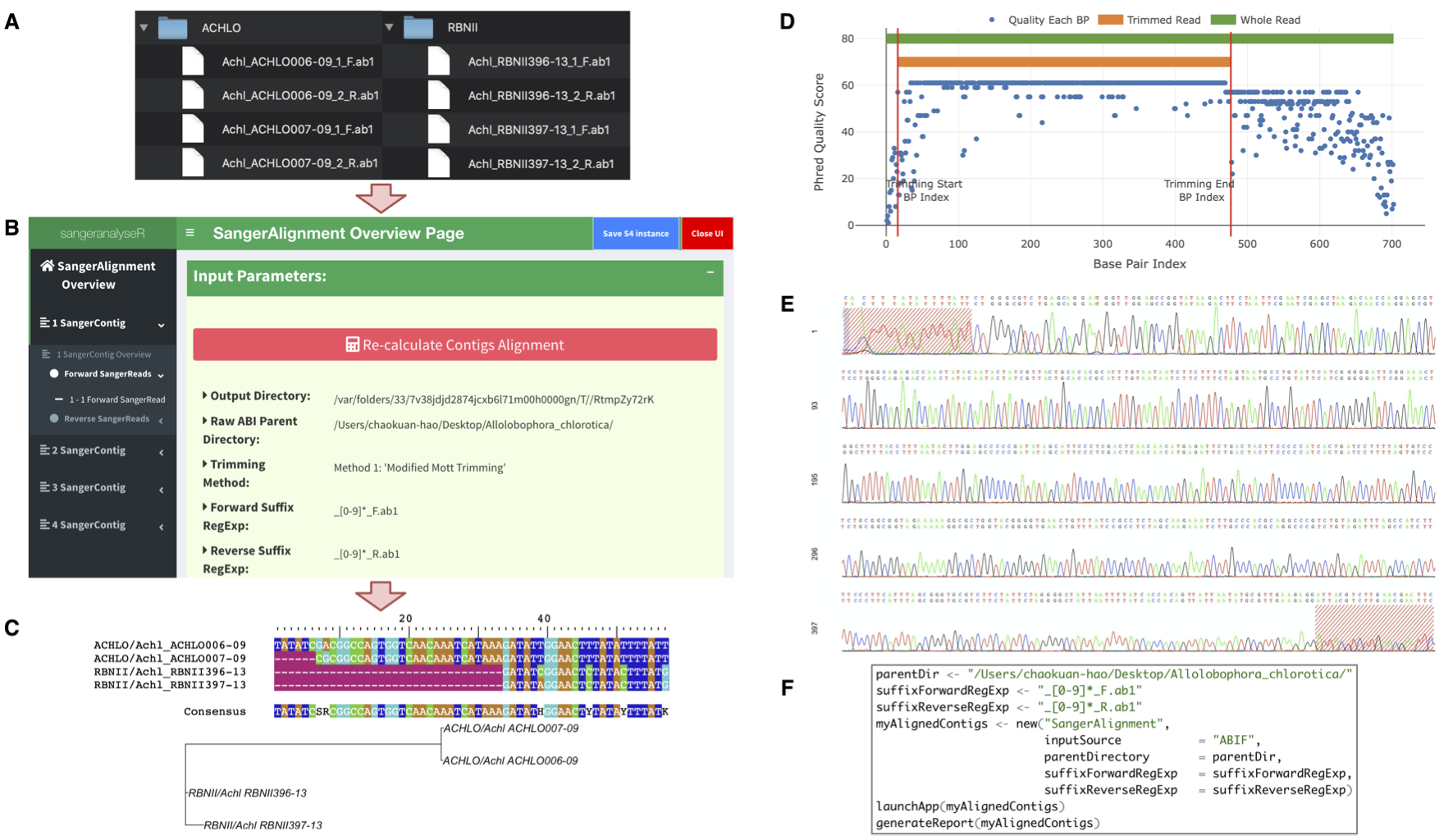
This package builds on sangerseqR to allow users to create contigs from collections of Sanger sequencing reads. It provides a wide range of options for a number of commonly-performed actions including read trimming, detecting secondary peaks, and detecting indels using a reference sequence. All parameters can be adjusted interactively either in R or in the associated Shiny applications. There is extensive online documentation, and the package can outputs detailed HTML reports, including chromatograms.
Source code & Documentation
sangeranalyseR: simple and interactive processing of Sanger sequencing data in R
Kuan-Hao Chao*, K. Barton, S. Palmer, and R. Lanfear* (2021). sangeranalyseR: simple and interactive processing of Sanger sequencing data in R, Genome Biology and Evolution, Volume 13, Issue 3, March 2021, evab028, https://doi.org/10.1093/gbe/evab028
[Bioc 2021] Bioconductor Conference 2021
Poster at Bioconductor Conference 2021 (Bioc 2021), Virtual
